Peptides, Polypeptides & Proteins
This lesson covers:
- How proteins are made of amino acids joined by peptide links
- Hydrolysis of polypeptides
- The three levels of protein structure
- The types of bonds that help maintain protein structure
- Separating amino acids by thin-layer chromatography
Proteins are polymers of amino acids
Proteins are composed of long chains of amino acids linked together.
- These chains of amino acids are created through condensation reactions between the amine group of one amino acid and the carboxyl group of another, with the release of a molecule of water.
- The bond that forms, known as a peptide bond, connects the amino acids into a polymer chain called a polypeptide.
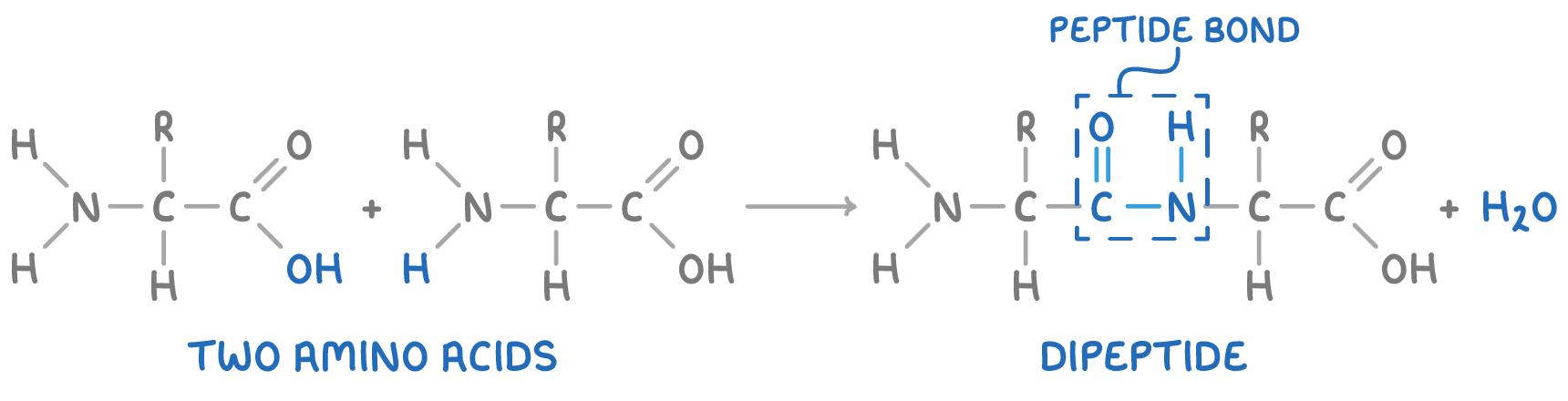
- A dipeptide is formed when two amino acids are joined by a peptide bond, whereas three linked amino acids create a tripeptide.
- When several amino acids join, they form small peptides, and longer chains are termed polypeptides or proteins.
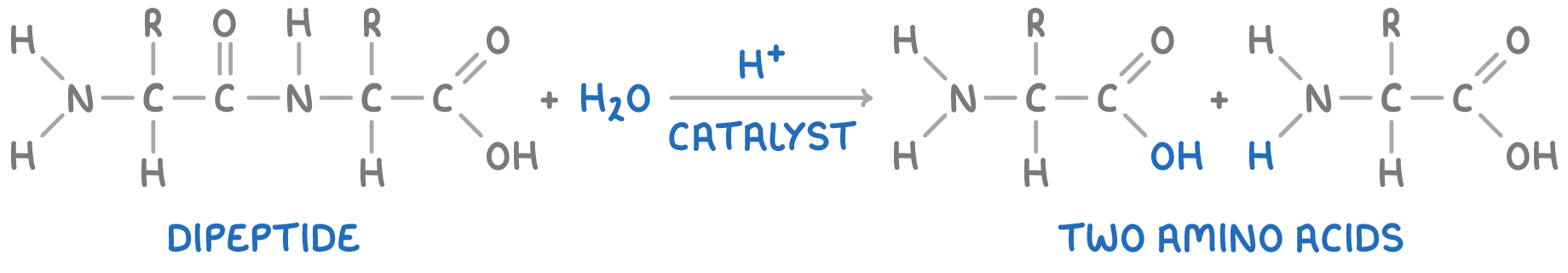
Hydrolysis of polypeptides
Polypeptide chains can be broken back down into amino acids through hydrolysis reactions, which involve the use of hot aqueous acid.
Proteins have three levels of structure
Proteins possess a complex three-dimensional structure that is established through three levels of organisation:
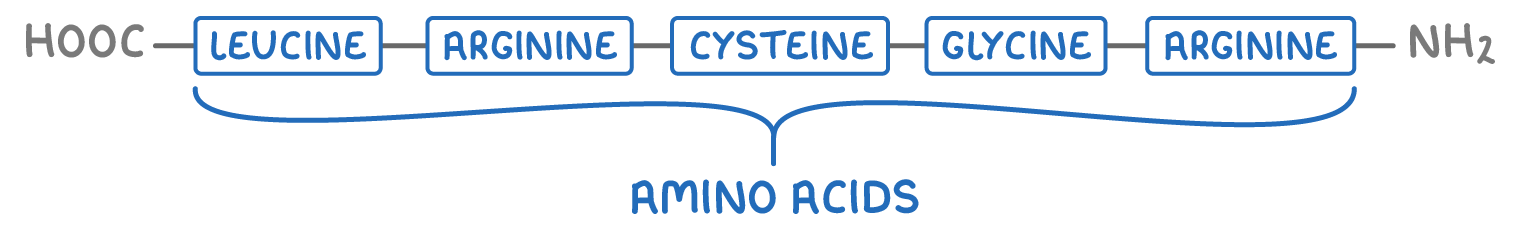
1. Primary structure - This is the sequence of amino acids in the polypeptide chain.
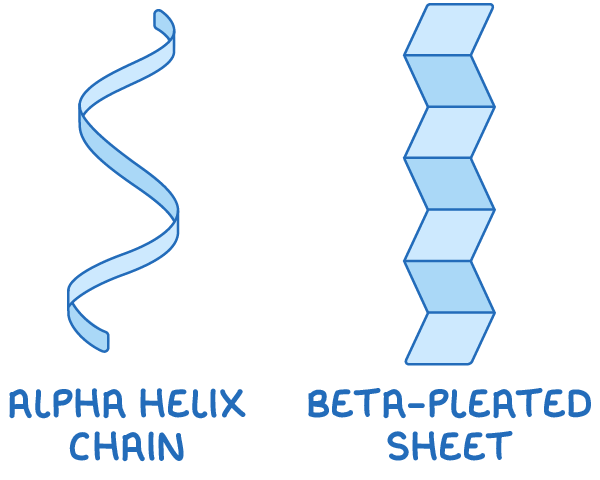
2. Secondary structure - This includes regular folding patterns like α-helices and β-pleated sheets, which are formed by hydrogen bonds between peptide bonds.
3. Tertiary structure - This refers to the overall 3D shape of the protein, encompassing its folding and coiling, which is maintained by interactions between R groups.
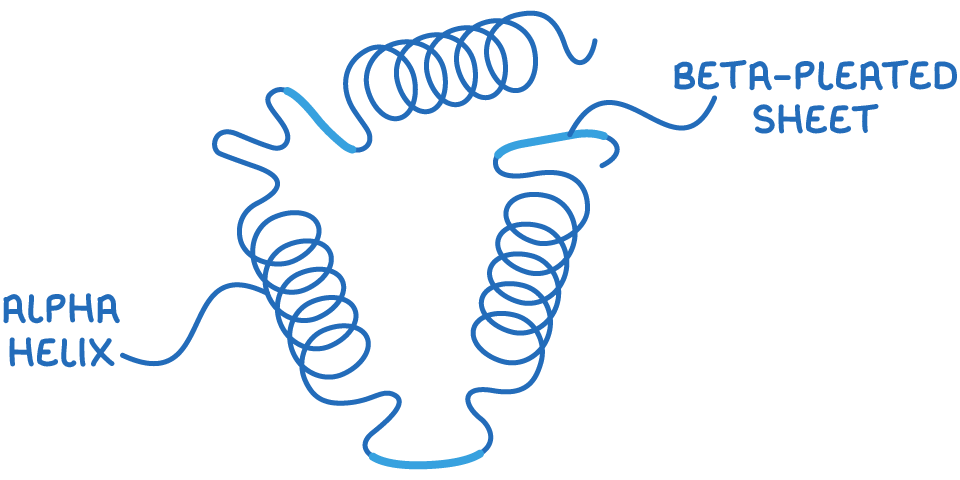
Bonds that maintain protein structure
The secondary and tertiary structures of proteins are held in place by two main types of bonds:
- Hydrogen bonds - These form between polar groups like -NH and -OH contribute to stabilising α-helices and β-pleated sheets.
- Disulfide bonds - These form between the -CH2SH side chains of cysteine amino acids, playing a crucial role in maintaining the protein's 3D structure.
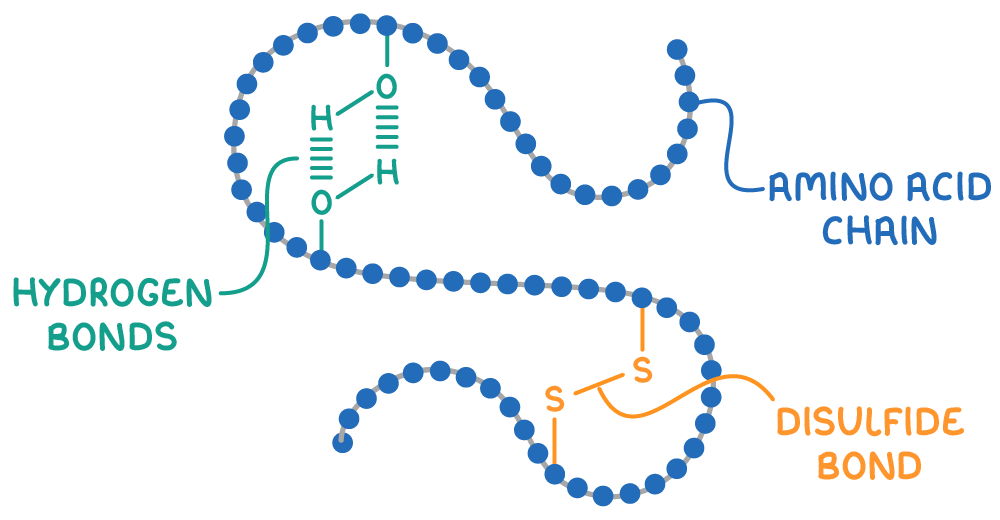
Alterations in temperature and pH can disrupt these bonds, leading to denaturation, a permanent change in the protein's shape that results in a loss of function.
Amino acid thin layer chromatography
Thin layer chromatography (TLC) is a technique used to separate and identify amino acids in a mixture.
The process involves the following steps:
- Sample application - The amino acid mixture is placed as a small spot on a TLC plate coated with a silica or alumina adsorbent, which serves as the stationary phase.
- Developing chamber - The plate is positioned in a developing chamber containing a solvent or solvent mixture, known as the mobile phase.
- Separation - As the solvent ascends the TLC plate by capillary action, it carries the amino acids upwards. The amino acids travel at different speeds based on their solubility in the solvent. Less soluble amino acids adhere more strongly to the silica, slowing their ascent, resulting in the separation of amino acids into distinct spots.
- Visualisation - Once the solvent has nearly reached the top of the plate, it is removed and dried. The amino acid spots are made visible using developing agents such as ninhydrin or ultraviolet light.
- Identification - The distance traveled by each amino acid spot relative to the solvent front is used to calculate the retention factor (Rf) values using the following equation:
Rf =distance moved by solventdistance moved by amino acid spot
By comparing the Rf values of the sample spots to known standards run on the same plate, the amino acids in the mixture can be identified.