Dihybrid Inheritance
This lesson covers:
- What a dihybrid cross is
- An example of dihybrid inheritance in pea plants
- Reasons for deviations from expected ratios
What is a dihybrid cross?
While monohybrid crosses show the inheritance of one gene with two alleles, dihybrid crosses show the simultaneous inheritance of two genes controlling separate characteristics.
The genotype in a dihybrid organism is written by placing the alleles for one gene immediately followed by the alleles for the second gene. It is important to keep the order consistent, always putting the same gene first to avoid confusion.
Dihybrid crosses can help to:
- Determine whether genes are linked.
- Locate genes on specific chromosomes.
- Calculate expected phenotypic ratios in subsequent generations.
Meiosis ensures that each gene still has just one allele in a dihybrid cross. The law of independent assortment means that alleles for different genes segregate independently during gamete formation in meiosis, unless they are linked.
An example of dihybrid inheritance in pea plants
Seed colour and seed texture in pea plants are determined by two different genes so are ideal for demonstrating dihybrid inheritance.
Dihybrid inheritance of seed colour and texture in pea plants can produce four different phenotypes:
| Phenotype | Genotypes |
|---|---|
| Yellow round seeds | YYRR |
| Green wrinkled seeds | yyrr |
| Yellow wrinkled seeds | YYrr or Yyrr |
| Green round seeds | yyRR or yyRr |
The F1 generation in true breeding pea plants
A true breeding or pure breeding organism is homozygous for the traits being studied. Selecting homozygous parents ensures predictable allele transmission during meiosis.
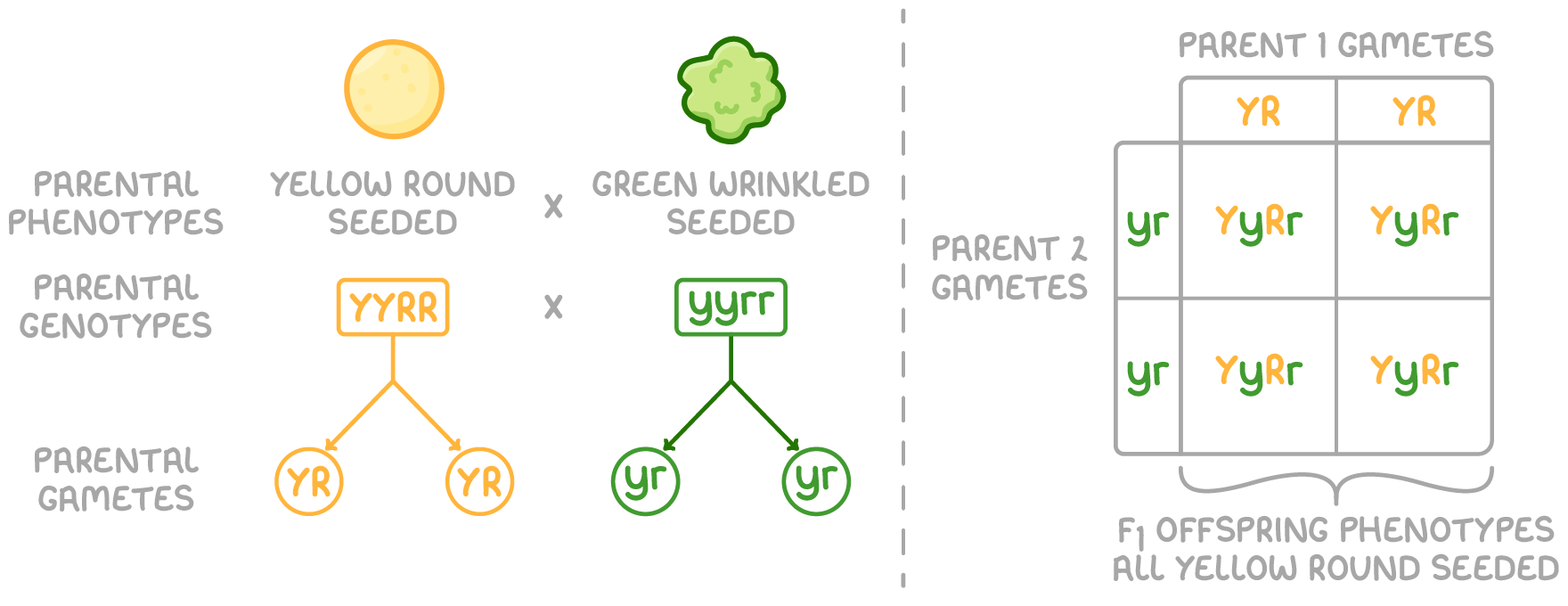
To set up a dihybrid cross for the F1 generation from true breeding pea plants:
- Mate two true breeding individuals with contrasting homozygous genotypes (YYRR and yyrr).
- Identify the alleles of the gametes formed during meiosis from these parents (YR and yr).
- Identify the F1 offspring genotypes from the random fusion of these parental gametes.
- Identify the F1 offspring phenotypes based on dominant and recessive alleles.
All F1 offspring have identical phenotypes and identical genotypes in a dihybrid cross between homozygotes. This shows that the two gene pairs have undergone independent assortment has occurred during meiosis, and indicates that the genes are not linked.
The F2 generation in true breeding pea plants
Crossing F1 individuals produces genetic variability in the form of new phenotypic combinations in the F2 offspring. Their phenotypic ratios depend on the dominance relationships at both gene loci.
For example, in the F2 offspring of yellow round F1 peas:
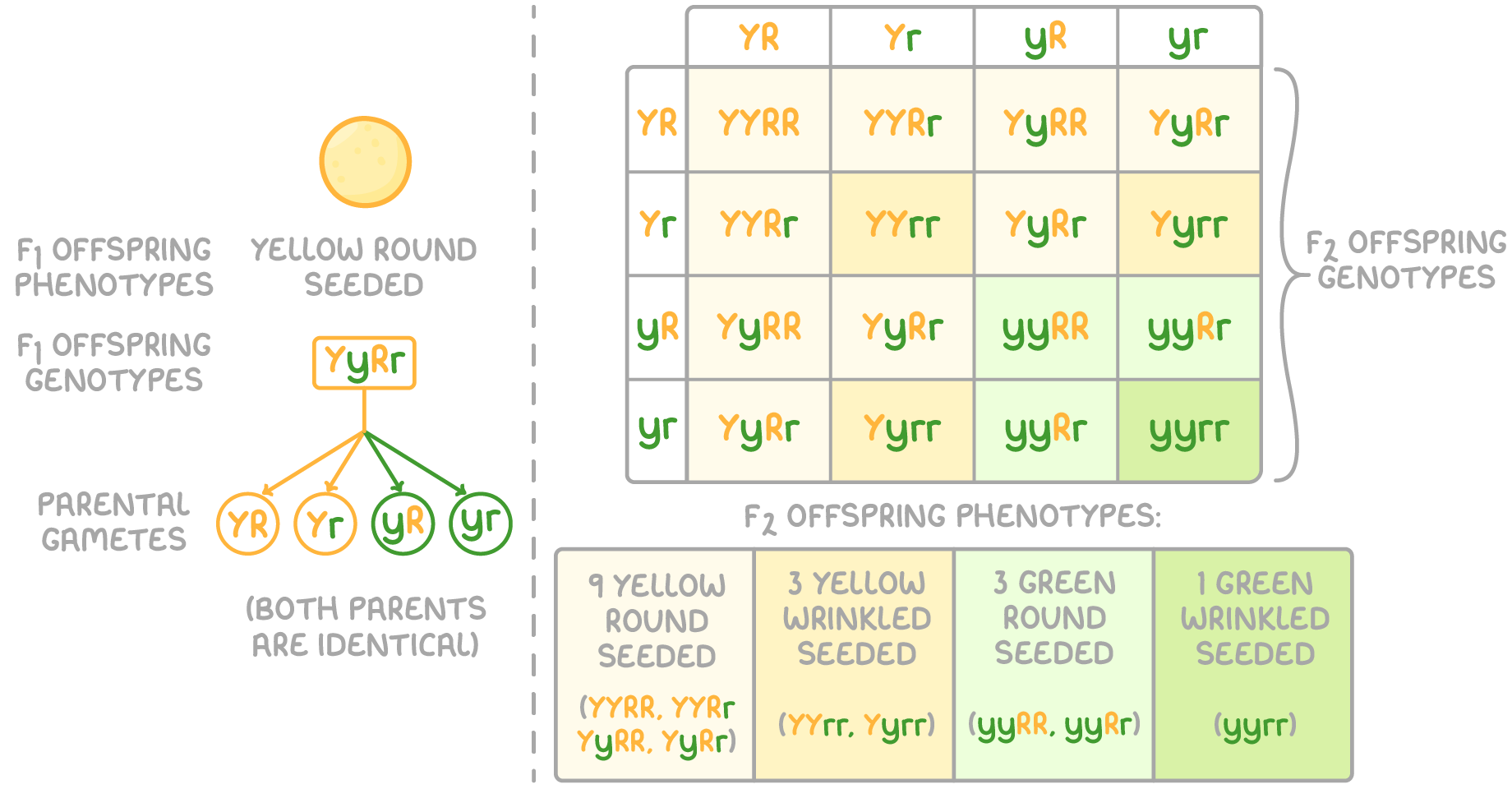
This 9:3:3:1 ratio illustrates the independent segregation of alleles controlling seed colour and texture. This ratio is typical of a dihybrid cross with both parents heterozygous at both gene loci.
Reasons for deviations from expected ratios
Dihybrid crosses predict specific phenotypic ratios, but actual observations can differ for two main reasons.
Random fertilisation:
- Gamete fusion is a chance process.
- Small sample sizes can lead to skewed ratios, while larger samples minimise these random effects.
Linked genes:
- Linked genes are on the same chromosome so alleles are usually inherited together, maintaining the parents’ original allele combinations in offspring.
- However, crossing over during meiosis can sometimes change these allele combinations by separating linked genes.