Producing DNA Fragments
This lesson covers:
- How DNA fragments can be transferred between organisms
- How reverse transcriptase can produce DNA fragments
- How restriction enzymes can produce DNA fragments
- How to synthesise custom DNA fragments
How DNA fragments can be transferred between organisms
Genetic engineering is the deliberate manipulation of genetic material to modify an organism's characteristics, often involving gene transfer.
Recombinant DNA is DNA that is altered to contain nucleotides from two different organisms. It allows DNA fragments to be transferred between organisms. Organisms that receive transferred DNA fragments are called genetically modified or transgenic organisms.
Key stages in gene transfer:
- The desired gene is identified and isolated.
- Multiple copies of the gene are made using the polymerase chain reaction (PCR).
- The gene is inserted into a vector.
- The vector delivers the gene into cells in a different organism.
- Cells with the new gene are identified, such as by using marker genes.
- Cells with the new gene are cloned.
Transgenic organisms can express the transferred gene to produce the encoded protein and express new genetic traits. This process is possible due to the universal nature of the genetic code and the similarity of transcription and translation across different organisms.
Methods of producing DNA fragments
Before transferring DNA, the target gene needs to be extracted.
There are three main methods used to produce DNA fragments for transfer:
- Making complementary DNA (cDNA) using reverse transcriptase and mRNA.
- Cleaving DNA from a donor organism with restriction enzymes.
- Synthesising new custom DNA sequences from nucleotides using a gene machine.
How reverse transcriptase can produce DNA fragments
Many cells contain lots of messenger RNA (mRNA) that corresponds to active genes. mRNA is easier to extract from cells than DNA. The enzyme reverse transcriptase can produce complementary DNA (cDNA) using mRNA templates to isolate target genes.
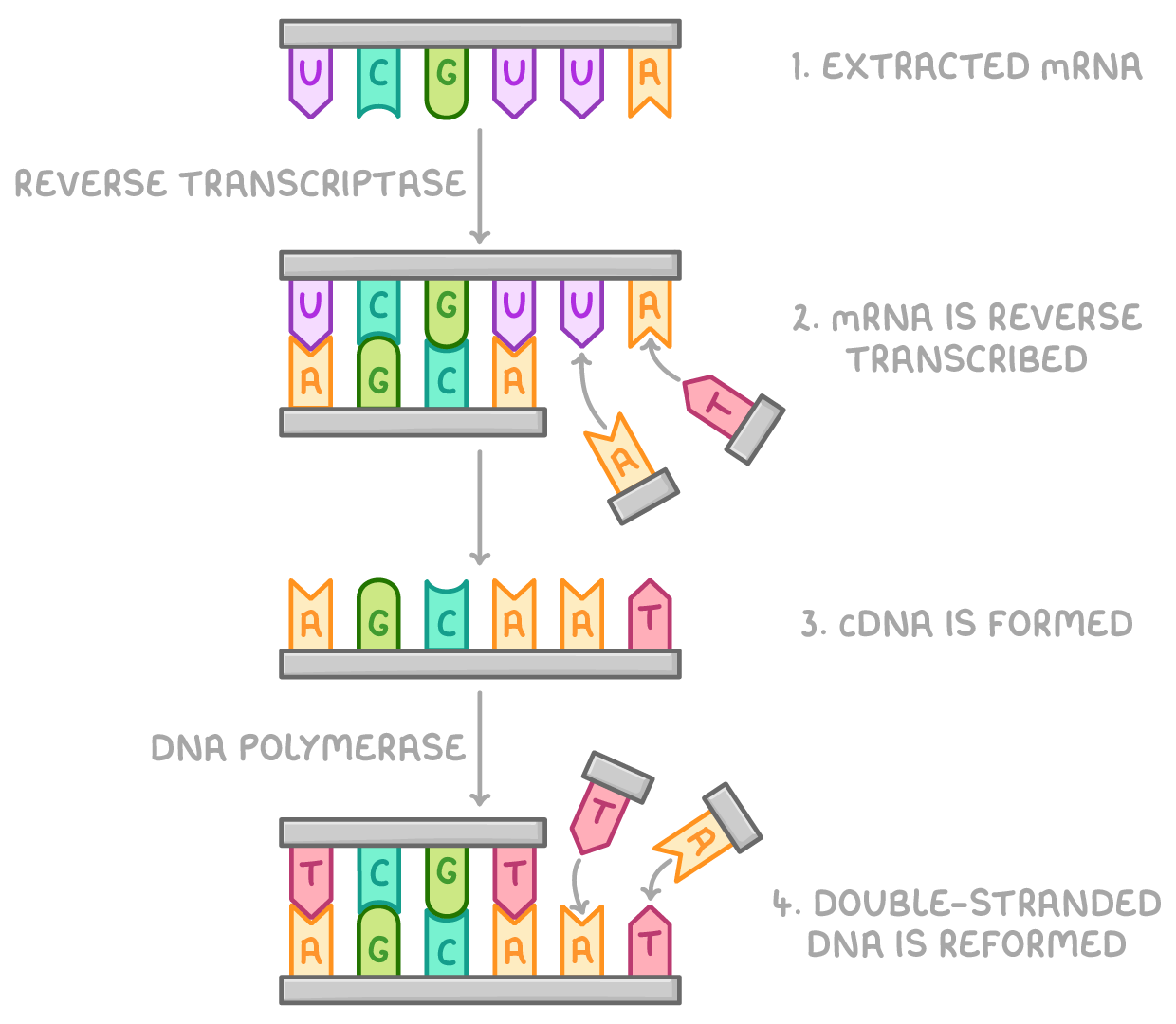
This process occurs as follows:
- mRNA is extracted from cells.
- mRNA is reverse transcribed using the reverse transcriptase enzyme and DNA nucleotides.
- This makes a cDNA strand identical to the original DNA strand, and cDNA is isolated from the mRNA strand.
- cDNA, free nucleotides, and DNA polymerase can then be used to form the other strand of DNA, reforming the desired gene.
This gives more copies of the sequence of interest to work with than just the two gene copies in the genome.
Using restriction enzymes to cut DNA
Restriction enzymes (restriction endonucleases) recognise and cut DNA at specific palindromic nucleotide sequences to isolate gene fragments. Palindromic sequences are sections of DNA where the nucleotide sequence reads the same in opposite directions on antiparallel strands.
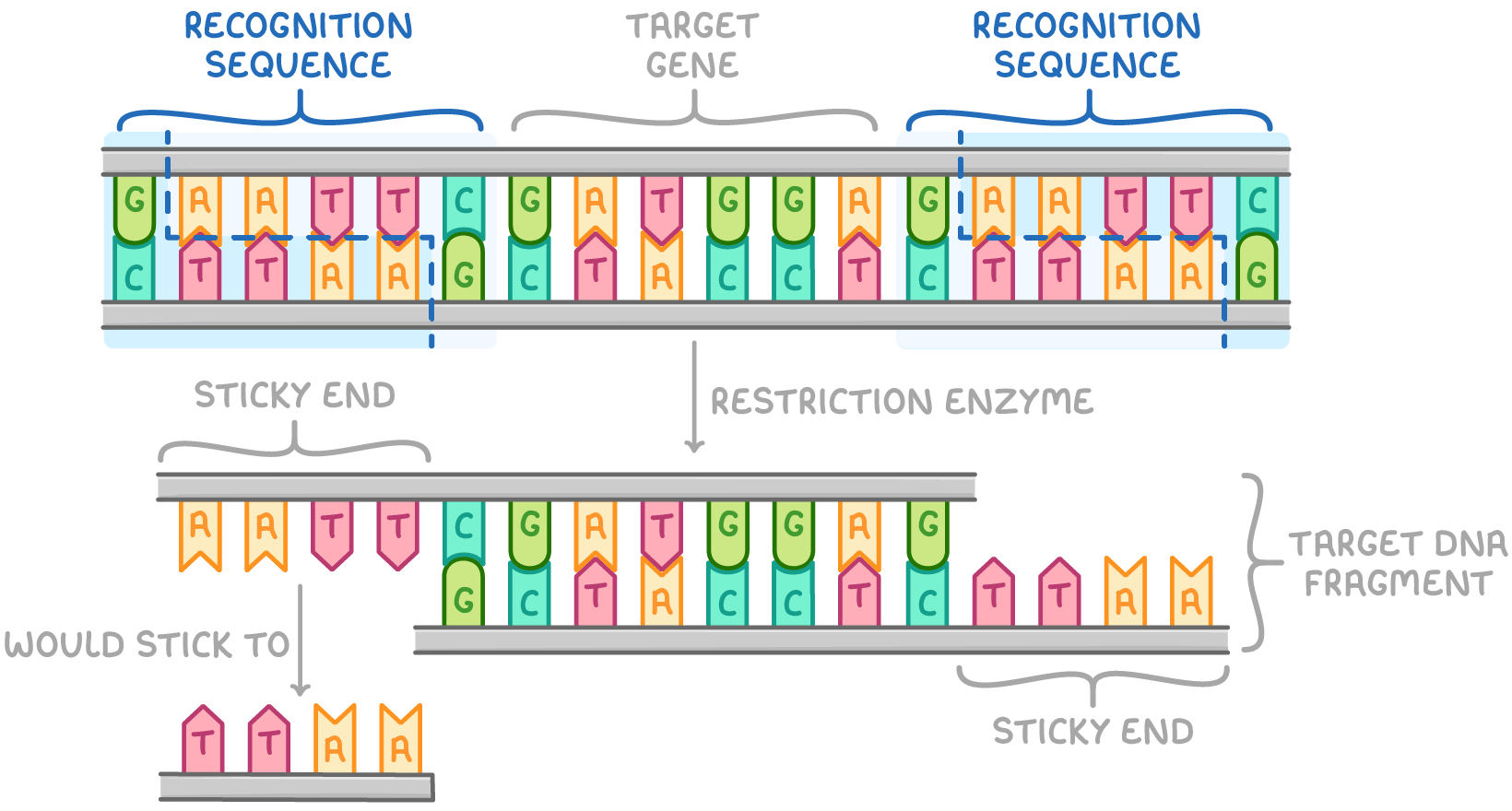
This process occurs as follows:
- DNA is incubated with chosen restriction enzyme(s).
- Restriction enzymes identify palindromic sequences in the DNA double helix and cut double-stranded DNA if their recognition sequence is present.
- Recognition sequences at either end of a desired DNA fragment allow restriction enzymes to separate the fragment from the rest of the DNA to obtain the desired gene.
- Enzymes cut target gene fragment out via hydrolysis reaction.
- Different restriction enzymes cut at different sequences based on their active site shape.
The cut DNA often has sticky ends. These are short overhanging sequences of unpaired bases that can bind to other DNA fragments when they are inserted into vectors.
Synthesising new custom DNA
Gene machines (DNA synthesisers) can now synthesise completely new DNA sequences.
This process occurs as follows:
- Choose codons for the desired amino acid sequence from a known protein structure.
- Use a computer to direct the synthesis of short fragments of DNA (oligonucleotides) in gene machines.
- Join the fragments together to make a longer sequence of nucleotides, forming the desired gene.
- Polymerase chain reaction (PCR) constructs a complementary DNA strand and amplifies the gene to produce multiple copies.
This method produces custom DNA without needing a natural template.