In Vivo Cloning
This lesson covers:
- Inserting a DNA fragment into a vector
- Using vectors to transfer DNA into host cells
- Identifying transformed host cells
- The need for promoter and terminator regions
Step 1 - Inserting DNA fragments into vectors
The process of producing large quantities of a target DNA fragment in living cells is sometimes called 'in vivo cloning' or 'in vivo DNA amplification'. It begins with inserting a target DNA fragment into a vector. This vector is then capable of transferring the DNA into a host cell.
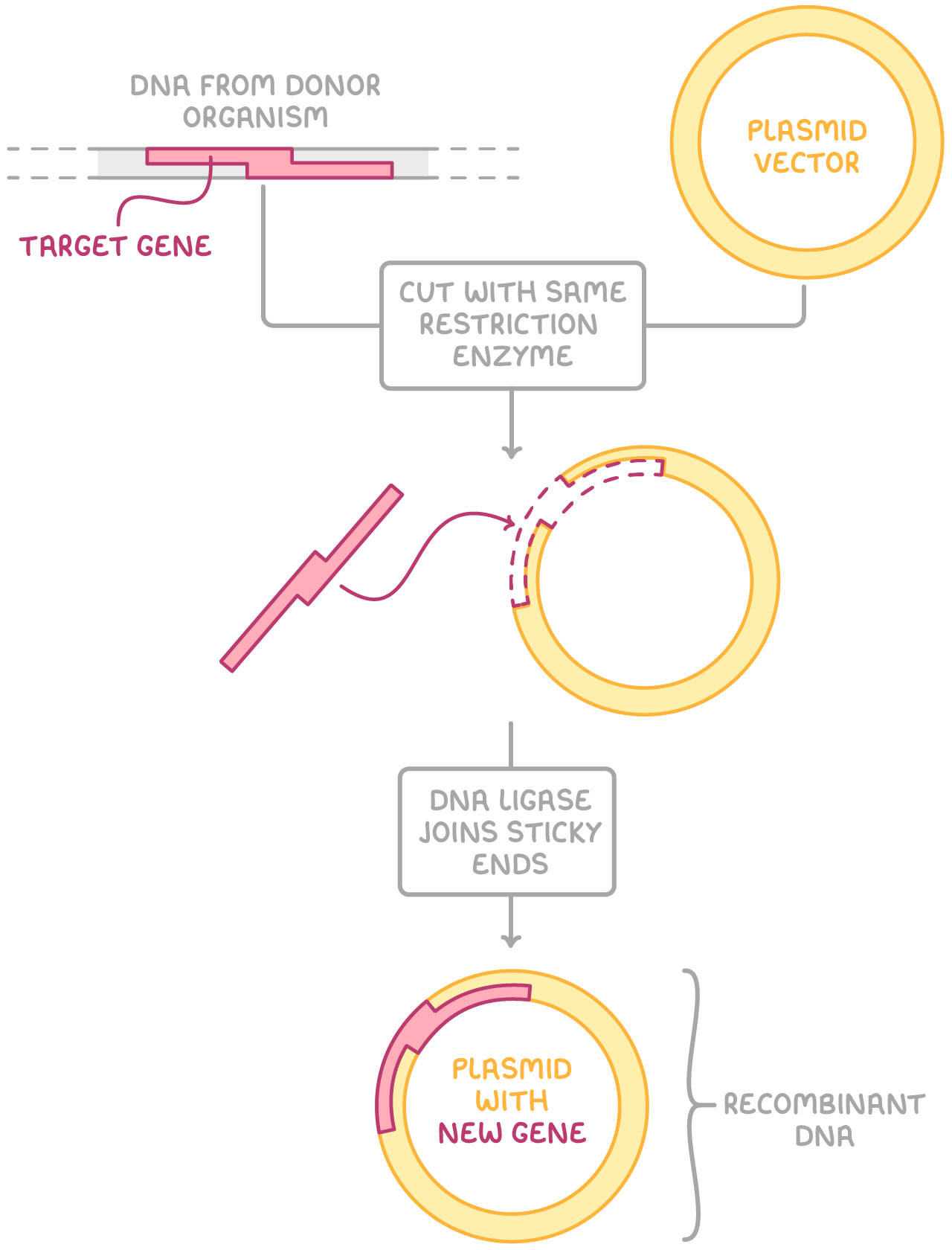
The key steps are:
- A vector is cut open at a specific site using a restriction enzyme, creating sticky ends.
- The same restriction enzyme is used to cut the target DNA fragment, creating complementary sticky ends.
- DNA ligase forms phosphodiester bonds between the sugar and phosphate groups on the two strands of DNA, joining the sticky ends of the vector and DNA fragment together.
- The newly formed combined DNA molecule is known as recombinant DNA.
Step 2 - Transferring recombinant DNA into host cells
Transformation involves introducing vectors with recombinant DNA into host cells, transforming these cells. The vectors are usually either plasmids or bacteriophages.
Plasmid vectors:
- These are small, circular DNA molecules, typically found in bacteria.
- Host cells are treated to enhance the uptake of plasmids that have recombinant DNA.
- For instance, applying calcium ions and temperature shifts can make bacterial membranes more permeable to plasmids.
Bacteriophage vectors:
- These are viruses that infect bacteria.
- Bacteriophages inject their DNA into host bacterial cells during infection.
- The phage DNA, now carrying the recombinant DNA, inserts into the host's DNA.
Step 3 - Identifying transformed host cells
As only around 5% of host cells may uptake DNA, it's important to identify which cells have been successfully transformed.
Marker genes indicate which host cells took up recombinant DNA:
- They are inserted into vectors alongside target genes.
- Transformed cells are cultivated on selective agar plates.
- Only transformed cells display the characteristics encoded by marker genes.
- These transformed cells can then be cultured to mass-produce the target DNA fragment through cellular replication.
Different types of marker genes
Different marker genes can identify which host cells took up recombinant DNA.
Some examples of marker genes include:
- A marker gene for a specific trait, like antibiotic resistance, ensures that only transformed cells form colonies.
- A marker gene that is visible under UV light like green fluorescent protein (GFP).
- Inserting a marker gene within the GFP gene inhibits fluorescence if it is successfully incorporated.
- A marker gene coding for an enzyme that alters the colour of a specific substrate.
Adding promoter and terminator regions
For the protein coded by an inserted DNA fragment to be produced, vectors require certain control elements.
Control elements that a vector needs:
- Promoter regions - Provide a binding area for RNA polymerase, signalling where the transcription of the gene should start.
- Terminator regions - Indicate where transcription should end, releasing RNA polymerase and terminating transcription.
Without these essential regions, the production of proteins from recombinant DNA would not occur. These promoter and terminator regions might already be present in the vector or may need to be added separately.